A Plot is Worth a Thousand Tests
Assessing Residual Diagnostics with the Lineup Protocol
2024-08-07
🔍 Regression Diagnostics
Diagnostics: is anything importantly wrong with my model?
\[\underbrace{\boldsymbol{e}}_\textrm{Residuals} = \underbrace{\boldsymbol{y}}_\textrm{Observations} - \underbrace{f(\boldsymbol{x})}_\textrm{Fitted values}\]
Residuals: what the regression model does not capture.
Checked by:
- Numerical summaries: variance, skewness, quantiles
- Statistical tests: F-test, BP test
- Diagnostic plots: residual plots, Q-Q plots
Diagnostic Plots
Residual plots are usually revealing when the assumptions are violated. –Draper and Smith (1998), Belsley, Kuh, and Welsch (1980)
Graphical methods are easier to use. –Cook and Weisberg (1982)
Residual plots are more informative in most practical situations than the corresponding conventional hypothesis tests. –Montgomery and Peck (1982)
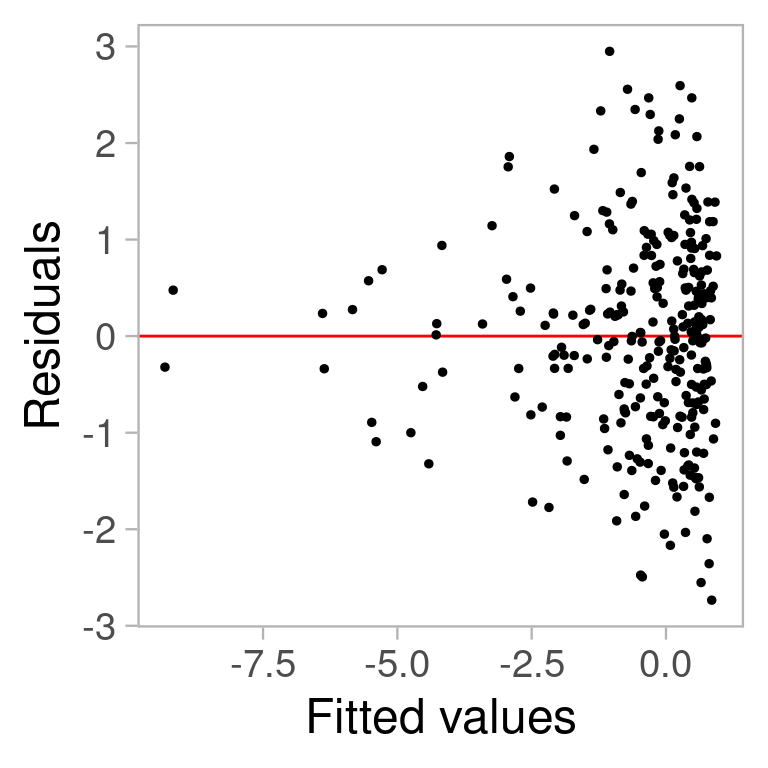
🤔 Plot Interpretation Challenges
What do you see?
- Vertical spread of the points varies with the fitted values.
=> heteroskedasticity?
- Triangle shape is actually from skewed distribution in x
Fitted model is fine!
We need an inferential framework to calibrate expectations when reading residual plots!
🔬 Visual Inference
Suggested by Buja et al. (2009)
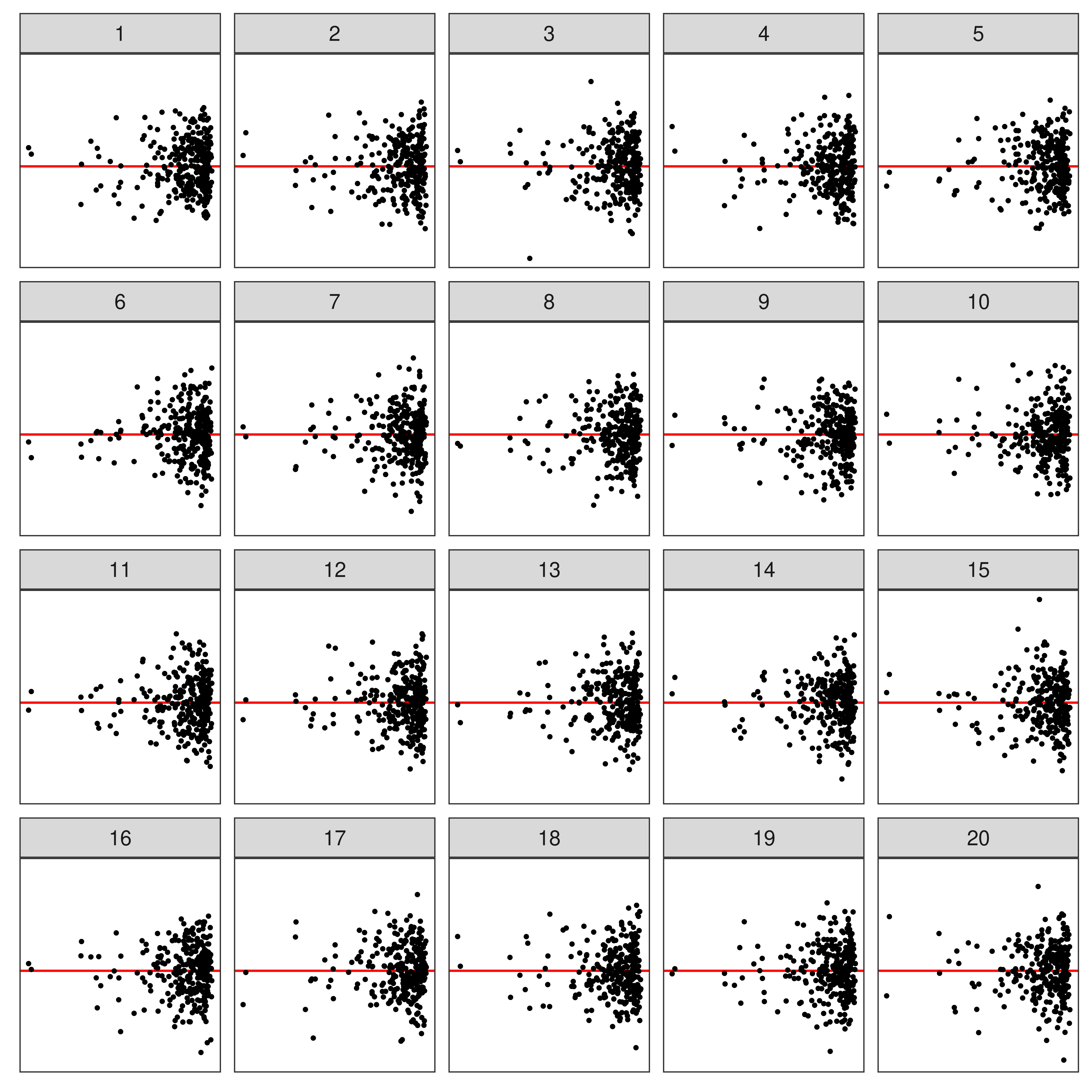
🔬 Visual Inference

Typically, a lineup of residual plots consists of
- 1 data plot
- 19 null plots w/ residuals simulated from the fitted model.
🔬 Visual Inference

To perform a visual test
- Observer(s) select the most different plot(s).
- P-value (“see value”) can be calculated via a beta-binomial model (VanderPlas et al. 2021)
🧪 Experiment
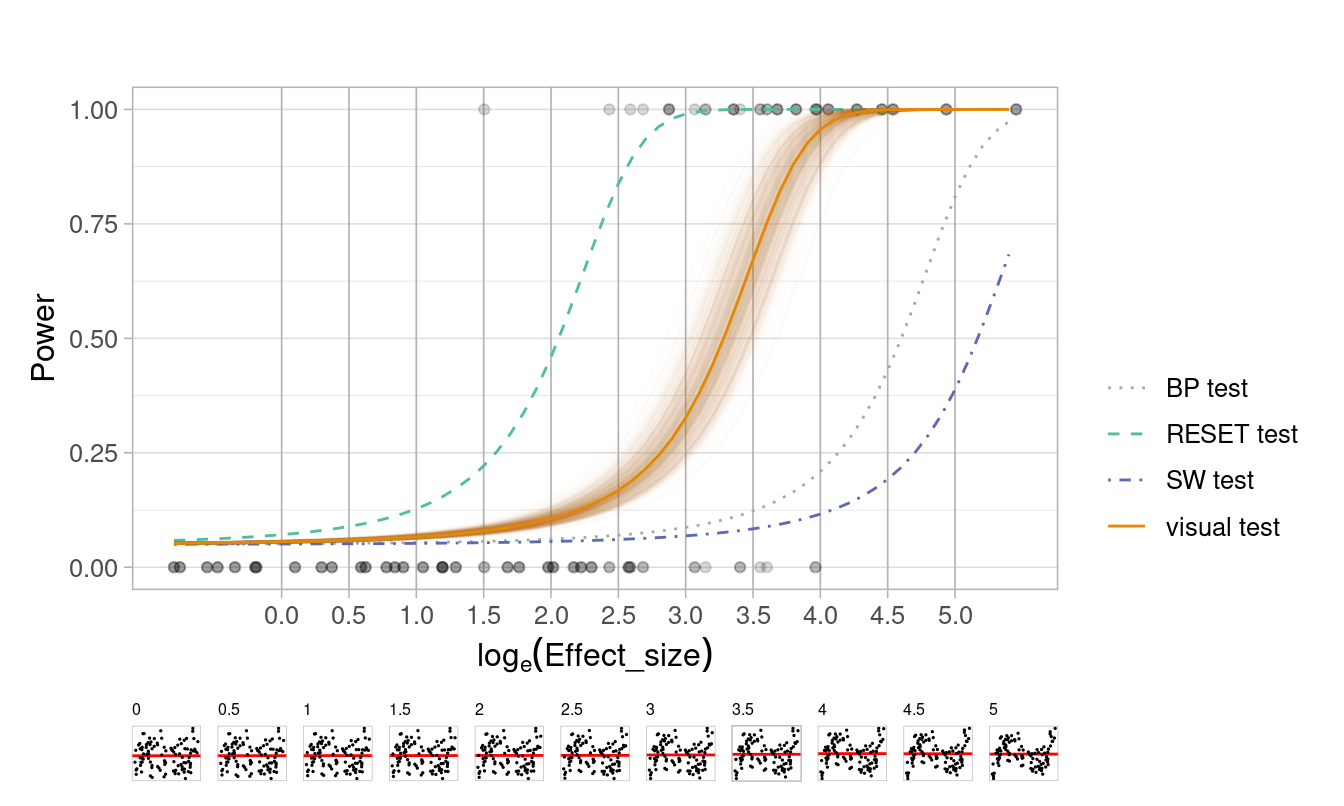
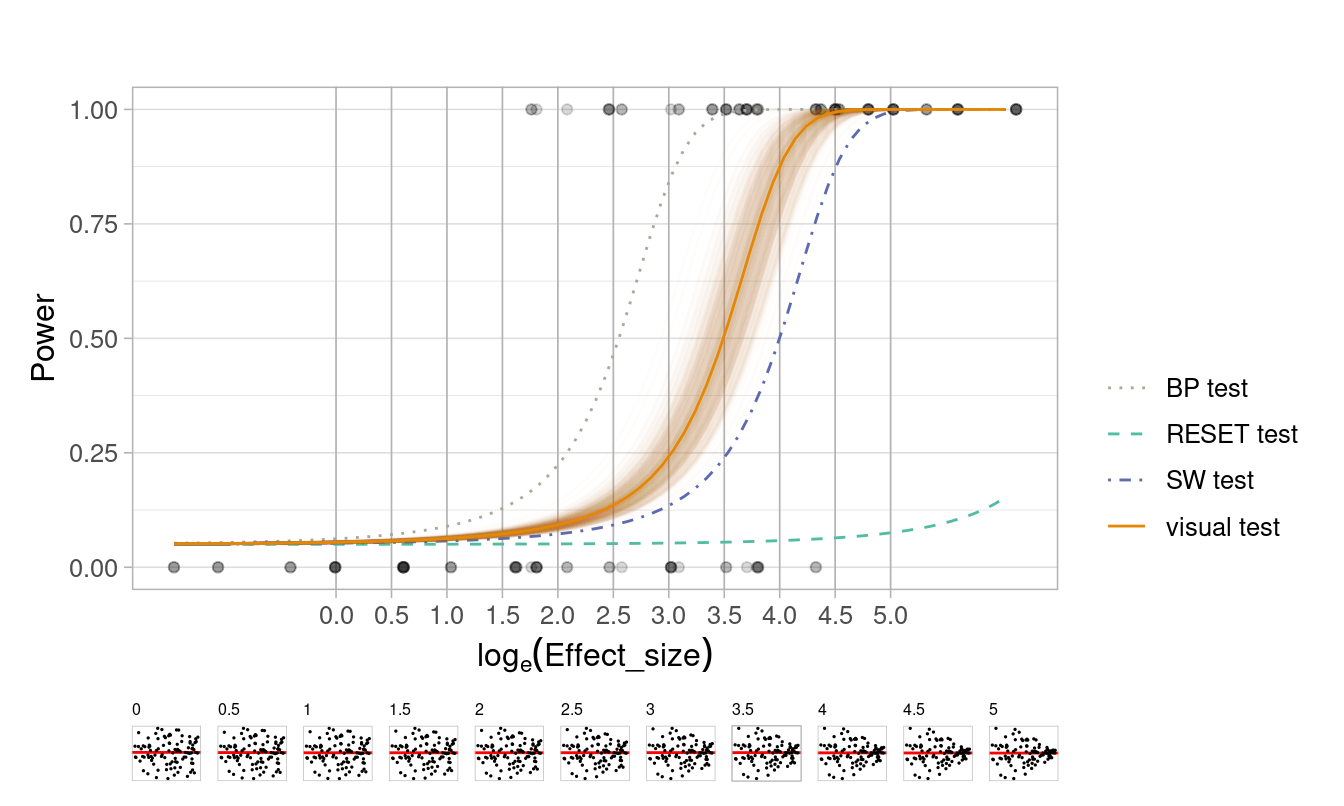
Compare conventional hypothesis testing with visual testing when evaluating residual plots
🖊 Design
| Model | Structure |
|---|---|
| Null | \(\boldsymbol{y} = \beta_0 + \beta_1\boldsymbol{x} + \boldsymbol{\varepsilon}\) |
| Non-linearity | \(\boldsymbol{y} = \boldsymbol{1} + \boldsymbol{x} + \boldsymbol{z} + \boldsymbol{\varepsilon}\) |
| Heteroskedasticity | \(\boldsymbol{y} = 1 + \boldsymbol{x} + \boldsymbol{\varepsilon}_h\) |
where
- \(\boldsymbol{\varepsilon} \sim N(\boldsymbol{0}, \sigma^2\boldsymbol{I})\)
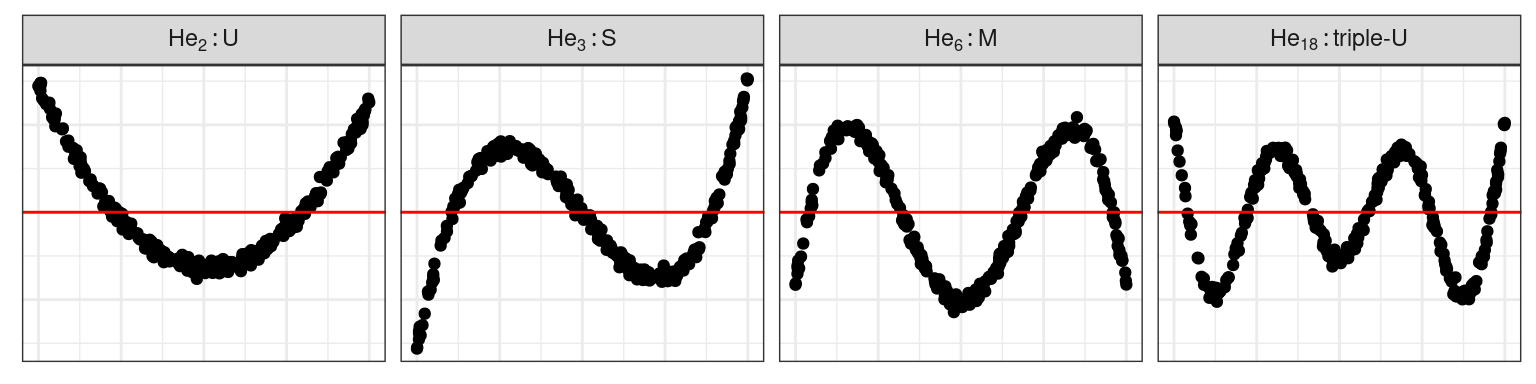
- \(\boldsymbol{z} \propto He_j(\boldsymbol{x})\), the \(j^{th}\) order probabilist Hermite polynomial
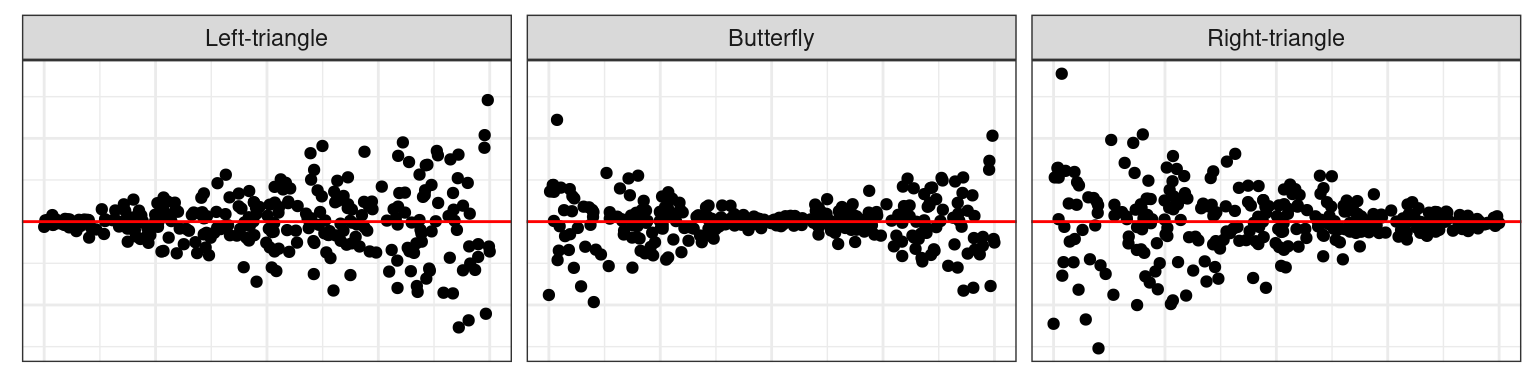
- \(\boldsymbol{\varepsilon_h} \sim N(\boldsymbol{0}, 1 + (2 - |a|)(\boldsymbol{x} - a)^2b \boldsymbol{I})\)
Non-linearity
Heteroskedasticity
Lineup Generation
parameters controlling signal strength (\(\sigma, b, n\))
\(4\times 4\times 3 \times 4 = 192\) non-linear parameter sets
\(3\times 5\times 3 \times 4 = 180\) heterosked. parameter sets
3 replicates per parameter set
576 (non linearity) + 540 (heteroskedasticity) lineups
(w/ \(\geq\) 5 evaluations\()\))36 Rorshach lineups to estimate \(\alpha\) for p-value calcs
📏 Effect size: Non-linearity
📏 Effect size: Heteroskedasticity
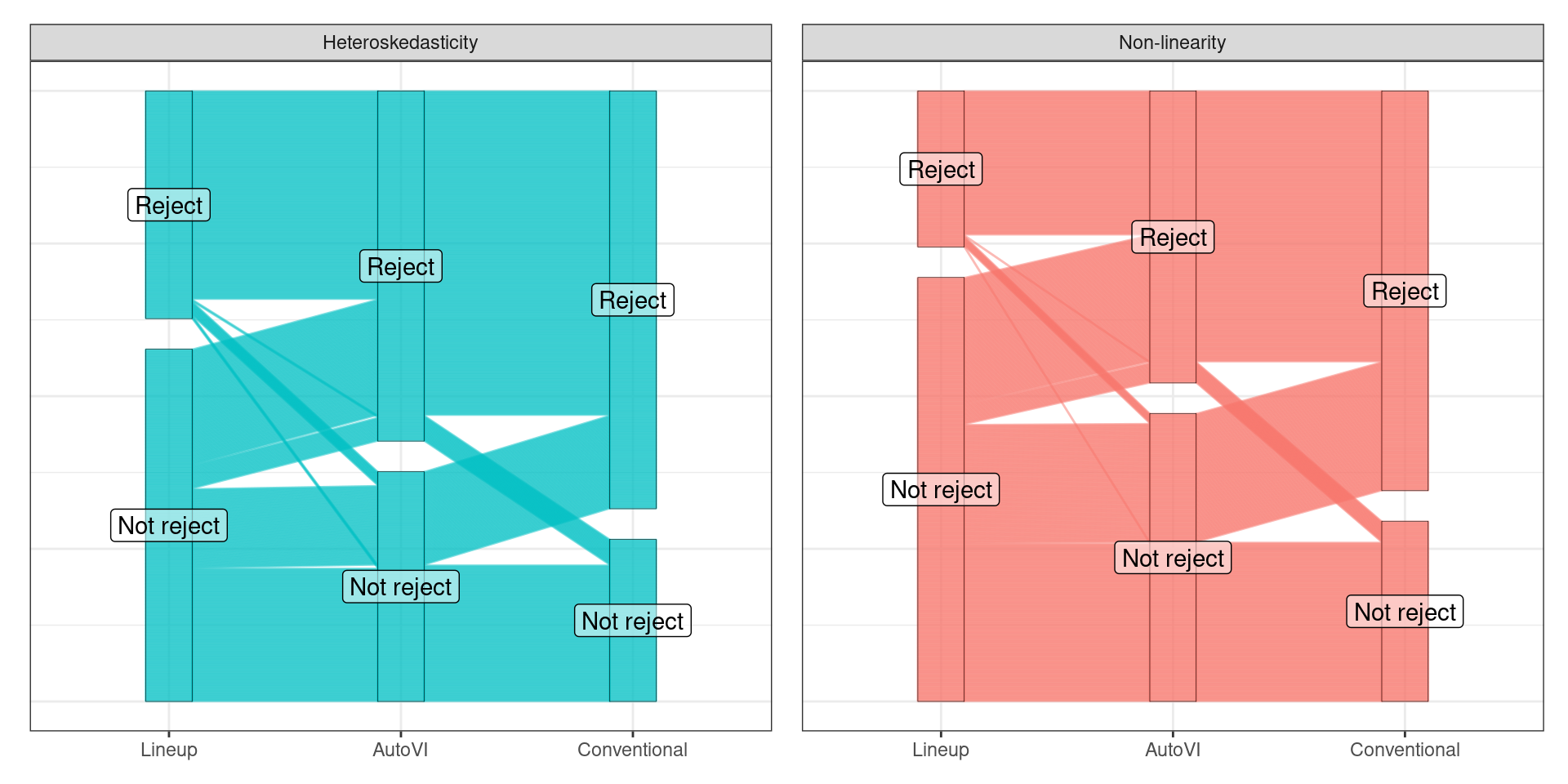
📊 Test Outcomes
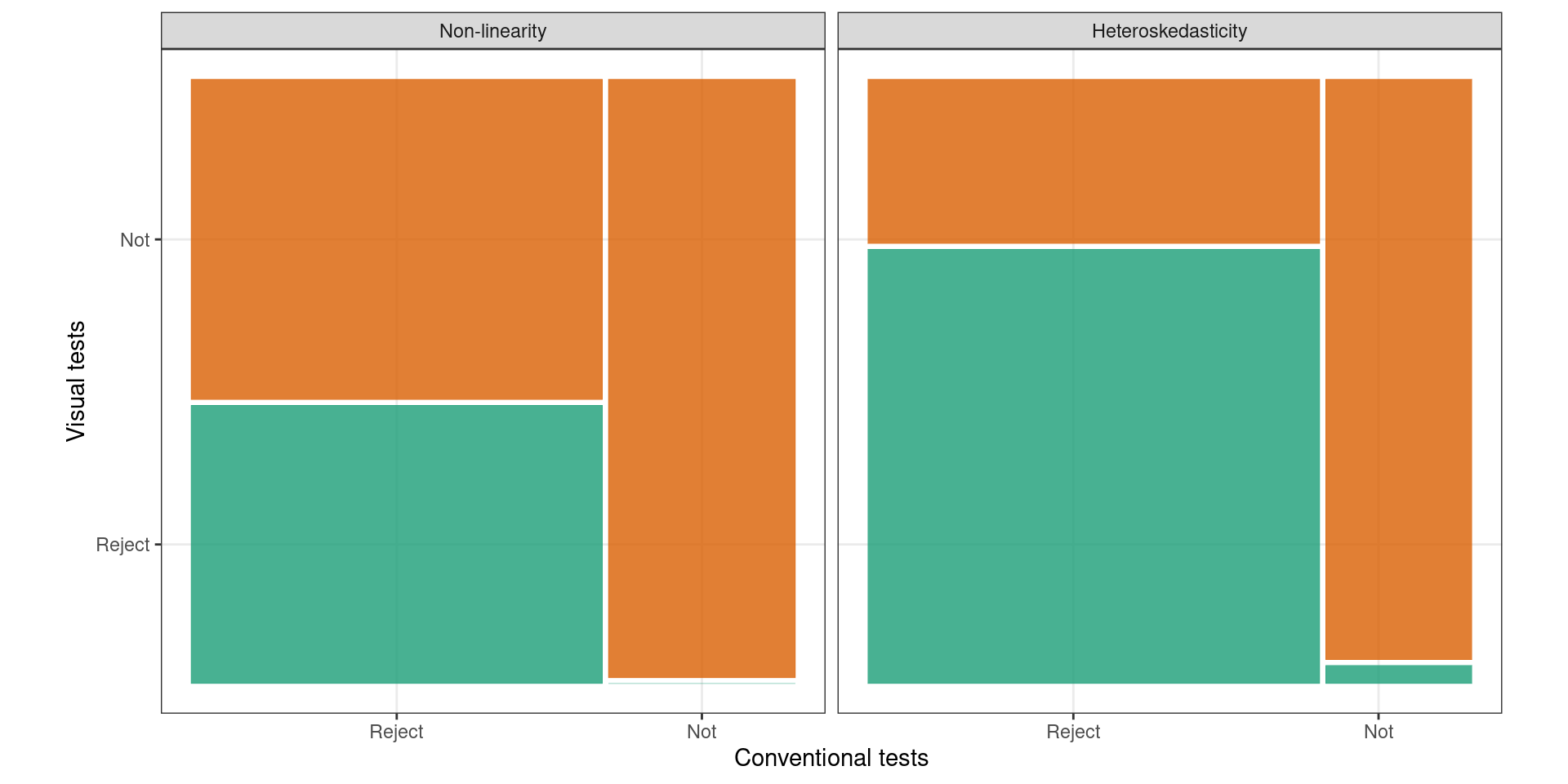
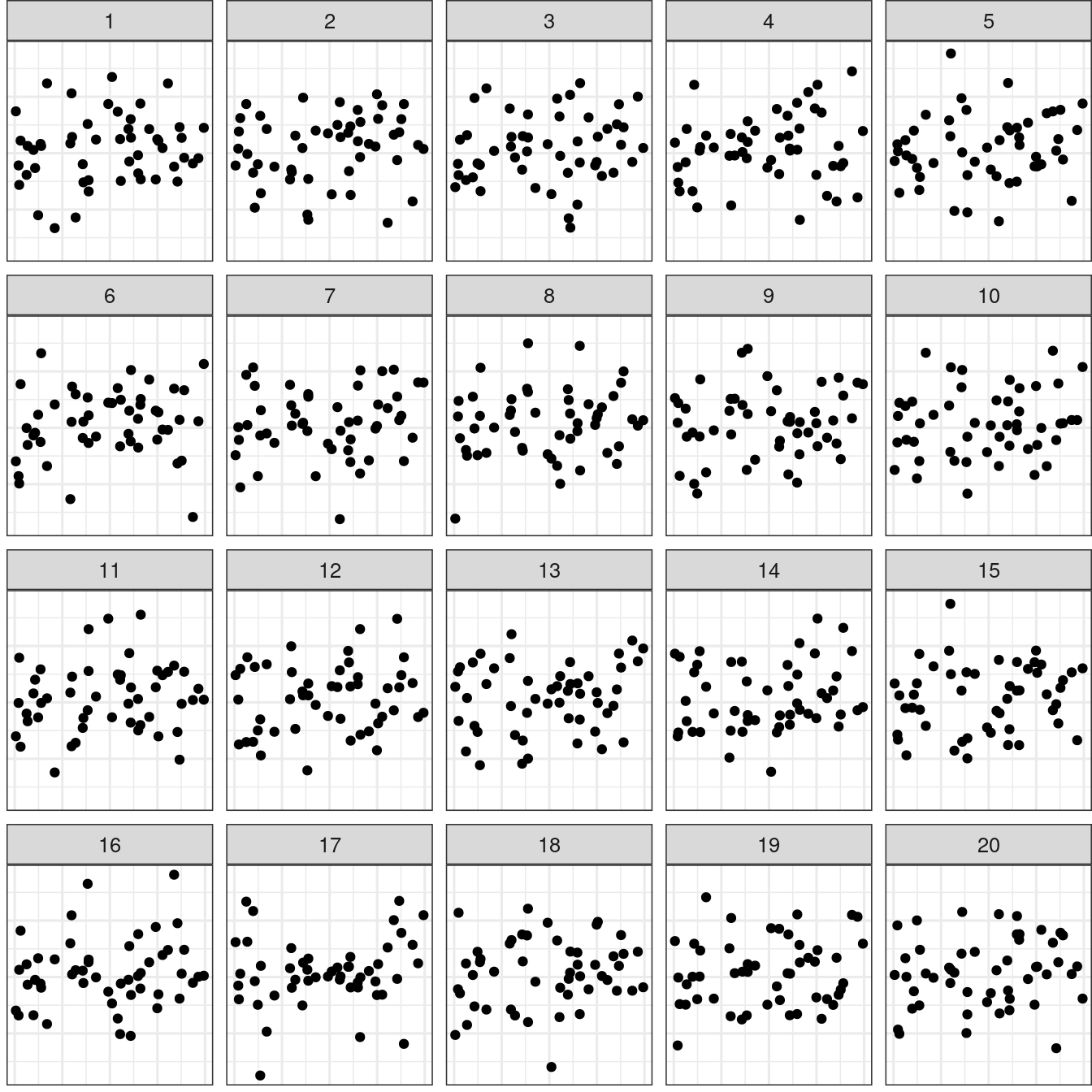
🪩 The Oddball Dataset
📓 Conclusions
Visual Test
- One test, multiple violations
Conventional Test
- Multiple tests required
| Violation | Test |
|---|---|
| nonlinearity | RESET |
| heteroskedasticity | Breusch-Pagan |
| goodness-of-fit | Shapiro-Wilk |
📓 Conclusions
Visual Test
- One test, multiple violations
- Reject ➡️
- severe issue w/ model fit
Conventional Test
- Multiple tests required
- Reject ➡️
- minor issue, no model impact OR
- major issue, model impact (no way to tell)
📓 Conclusions
Visual Test
- One test, multiple violations
- Reject ➡️
- severe issue w/ model fit
- 99.99% chance conventional test also rejects
Conventional Test
- Multiple tests required
- Reject ➡️
- minor issue, no model impact OR
- major issue, model impact (no way to tell)
⚠️ Limitations of Lineup Protocol
- Humans cannot (easily) evaluate
- lineups w/ many plots

- a large number of lineups
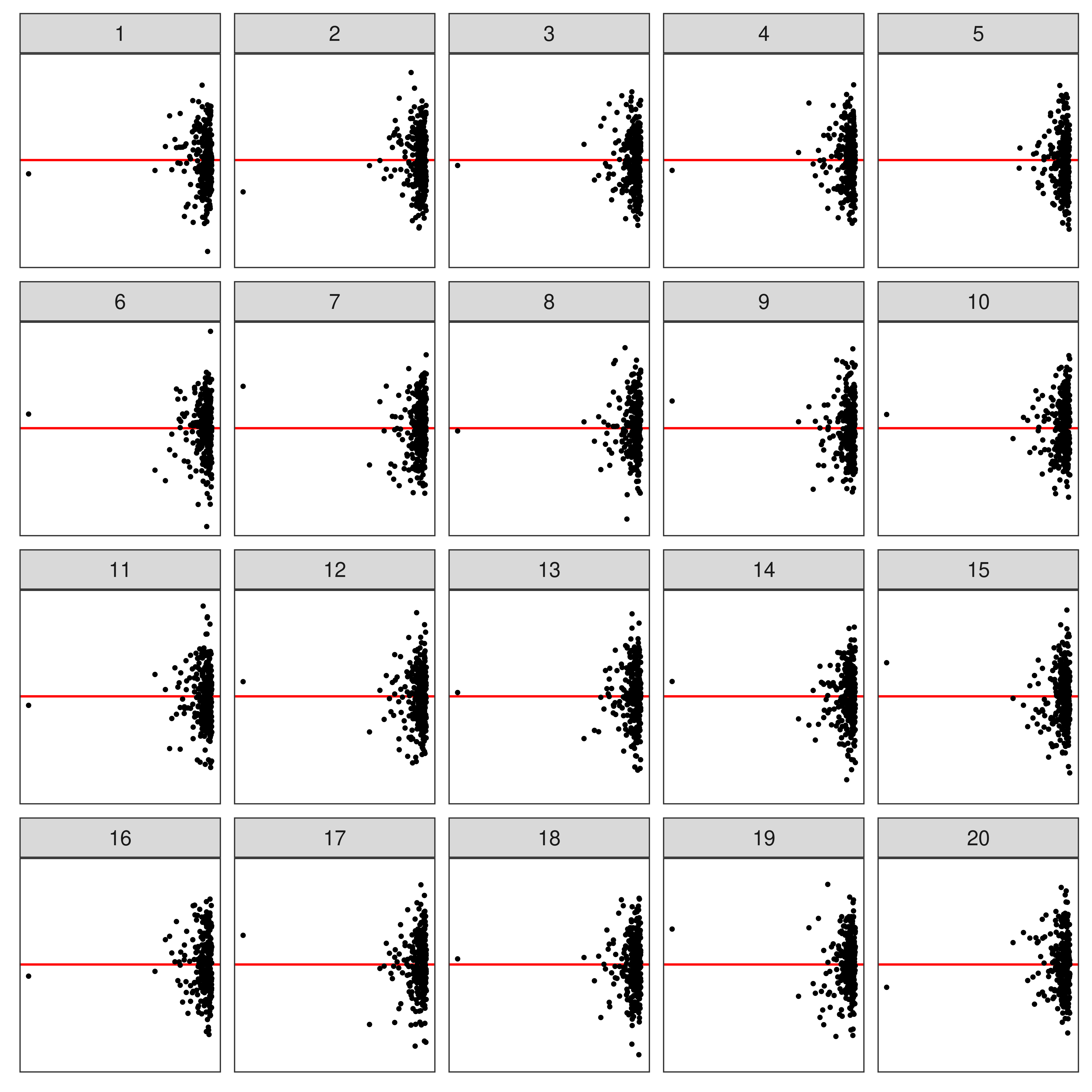
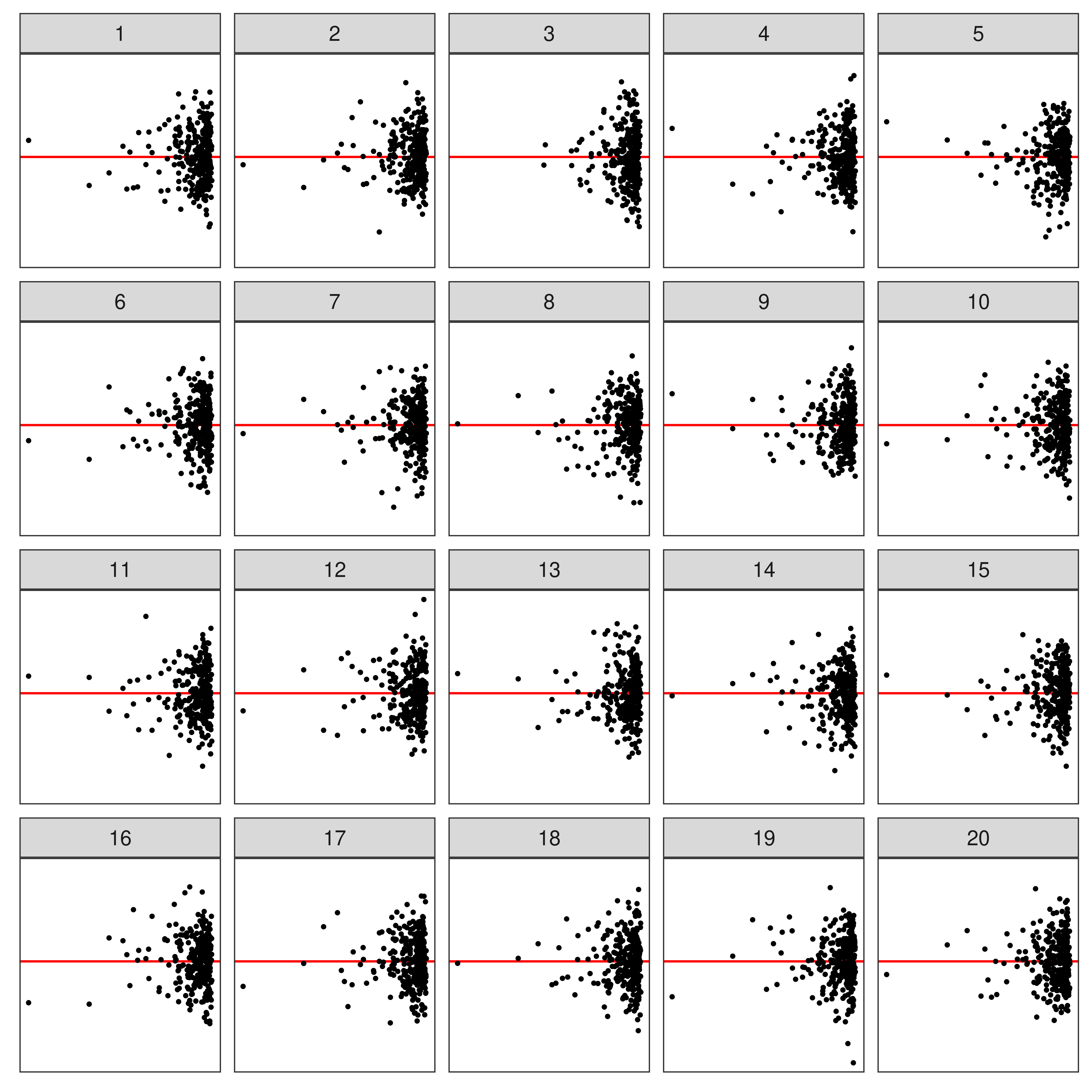
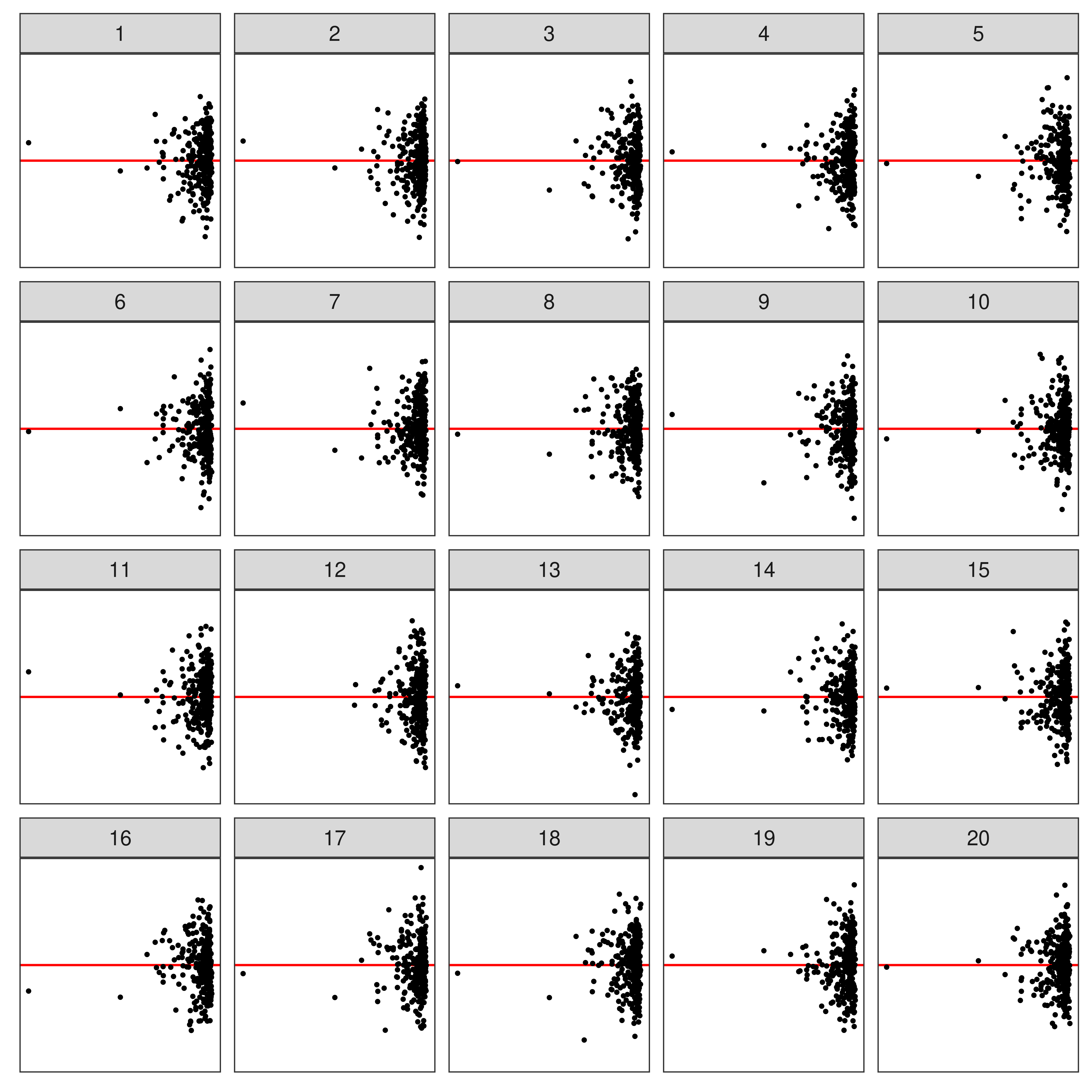
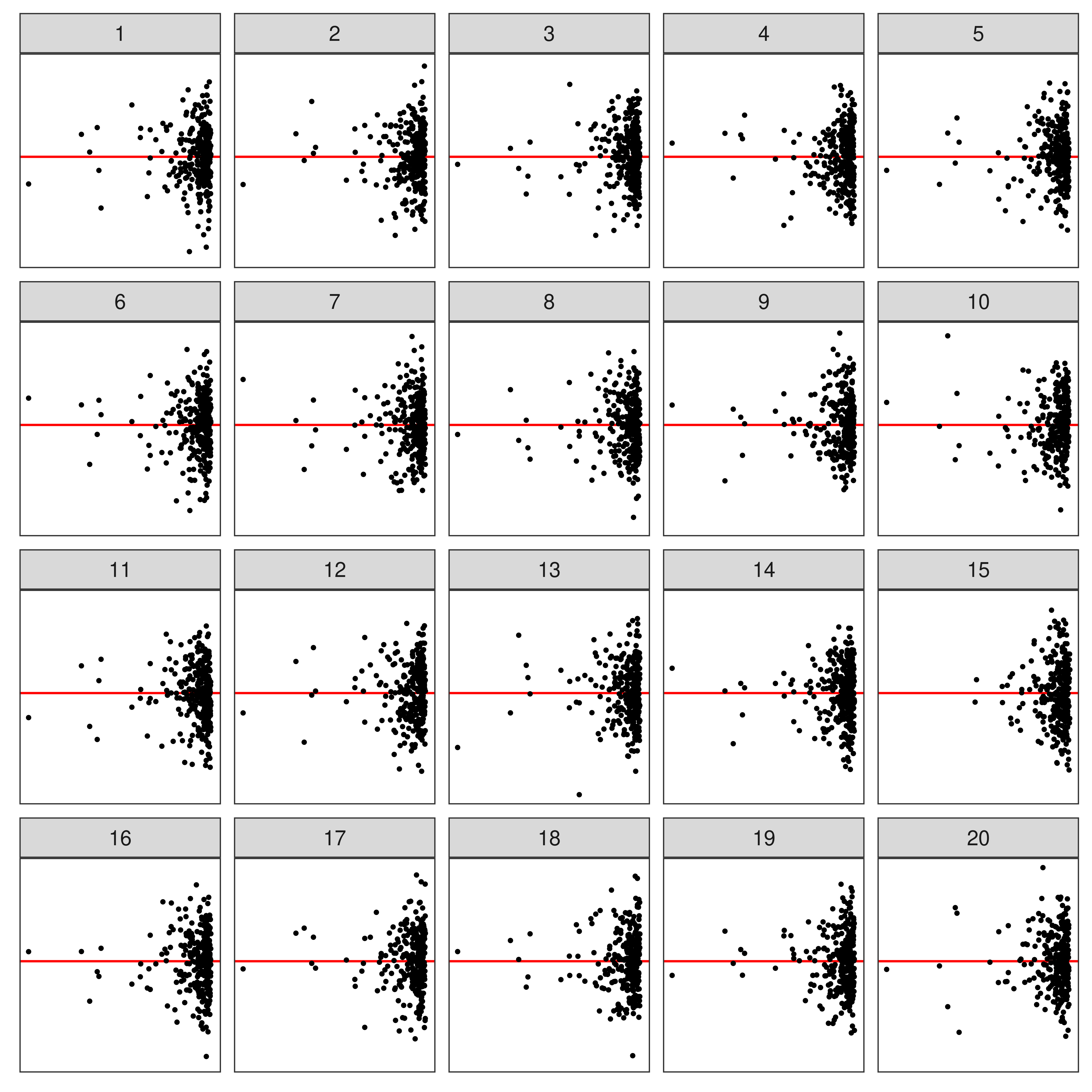
⚠️ Limitations of Lineup Protocol
- Humans cannot (easily) evaluate
- lineups w/ many plots
- a large number of lineups
- Lineups have 💰 high labor costs and can be 🕑 time consuming to evaluate
➡️ Make the 🖥️ do it for us with 🪄Computer Vision 🤖
🤖 AutoVI: Automated Assessment of Residual Plots with Computer Vision
🛣️ Roadmap
Estimate ‘visual’ distance \(D\) between
- an actual residual plot
- a plot of residuals generated under the null model
Compare \(\widehat D\) to a distribution of values
Calibrate against visual and conventional test results
📏 Measuring Distance
How to measure “difference”/“distance” between plots?
- Statistics: KL-Divergence (actual vs. null) when data generating process is known
- Graphics: scagnostics
- Image Analysis:
- pixel-wise sum of square differences
- Structural Similarity Index Measure (SSIM)
🎯 Estimating Distance
\(\widehat{D} = f_{CV}(V_{h \times w}(\boldsymbol{e}), n, S(\hat y, \hat e))\), where
- \(V_{h \times w}(.)\) is a \(h\times w\) image
- \(n\) observations
- \(S(\hat y, \hat e)\) are Scagnostics
- Computer Vision algorithm \(f_{CV}(.) \rightarrow [0, +\infty)\)
💡Training: Model Violations
Non-linearity + Heteroskedasticity
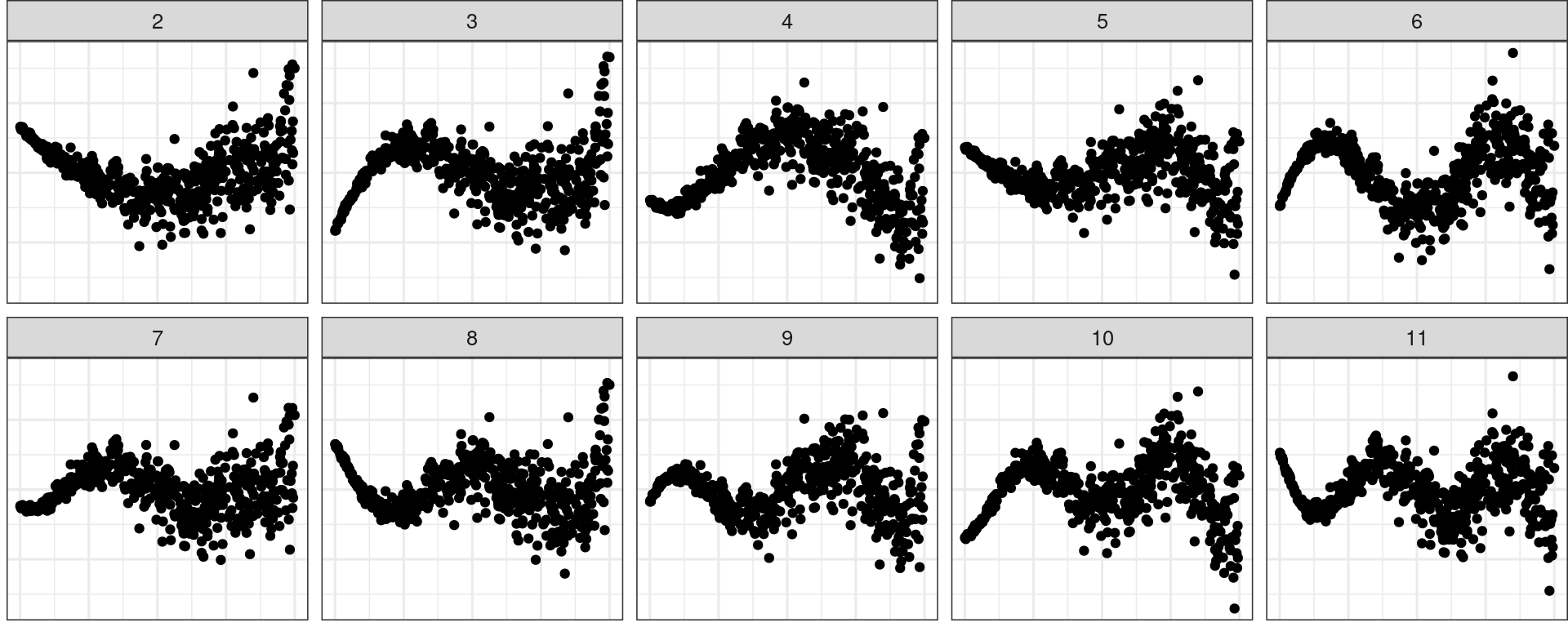
Non-normality + Heteroskedasticity
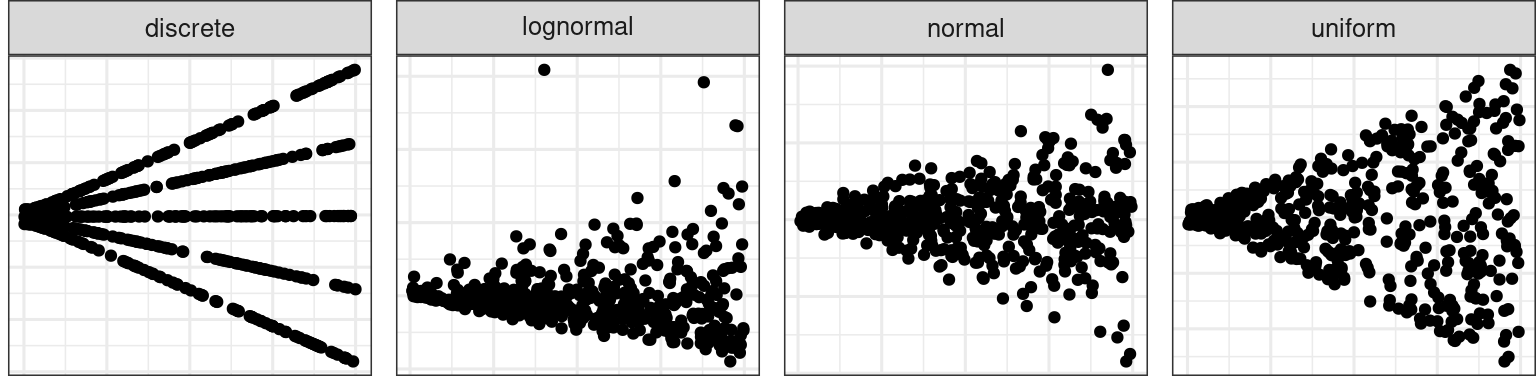
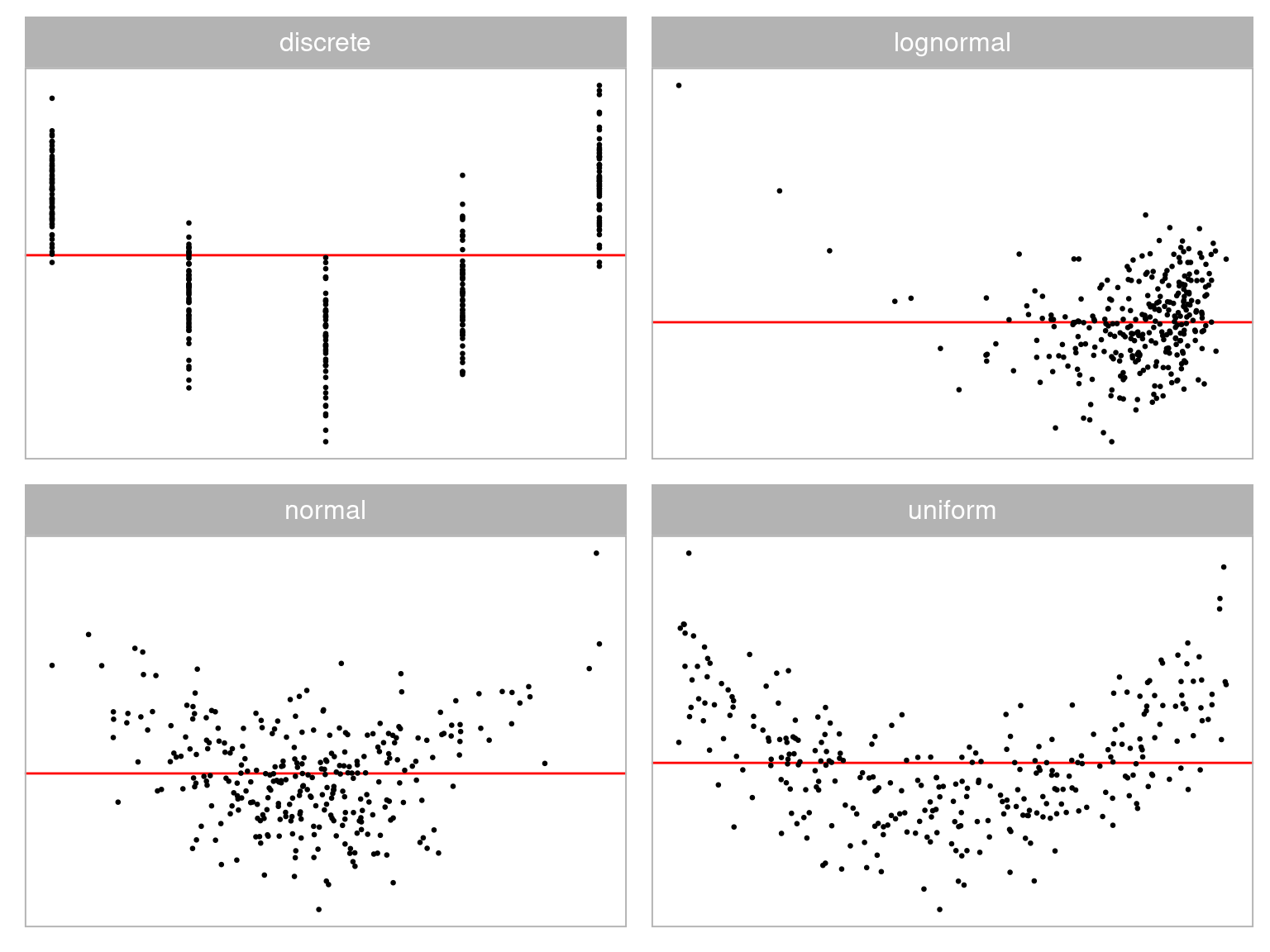
💡Training: Predictor Distribution
Distribution of predictor
🔬Statistical Testing
Estimate null distribution \(F(D | H_0)\) empirically:
- Generate data under \(H_0\)
- Calculate \(\widehat{D}\)
Compute critical value \(Q_0(0.95)\) as the value \(Q\) s.t. \(F(D \leq Q | H_0) = 0.95\)
\(p\)-value: \(P(D \geq D^\ast)\) for observed \(D^\ast\)
Comparison to Visual Inference
- Each lineup used in the experiment has 1 target and 19 null plots
- Run each plot through the model to get \(\widehat D\) (+ RESET & BP tests)
- Compare performance metrics
Comparison to Visual Inference
autovi Package
The autovi package provides automated visual inference with computer vision models. It is available on CRAN and Github.
Core Methods
- Null residuals simulation:
rotate_resid() - Visual signal strength:
vss() - Comprehensive checks:
check()andsummary_plot()
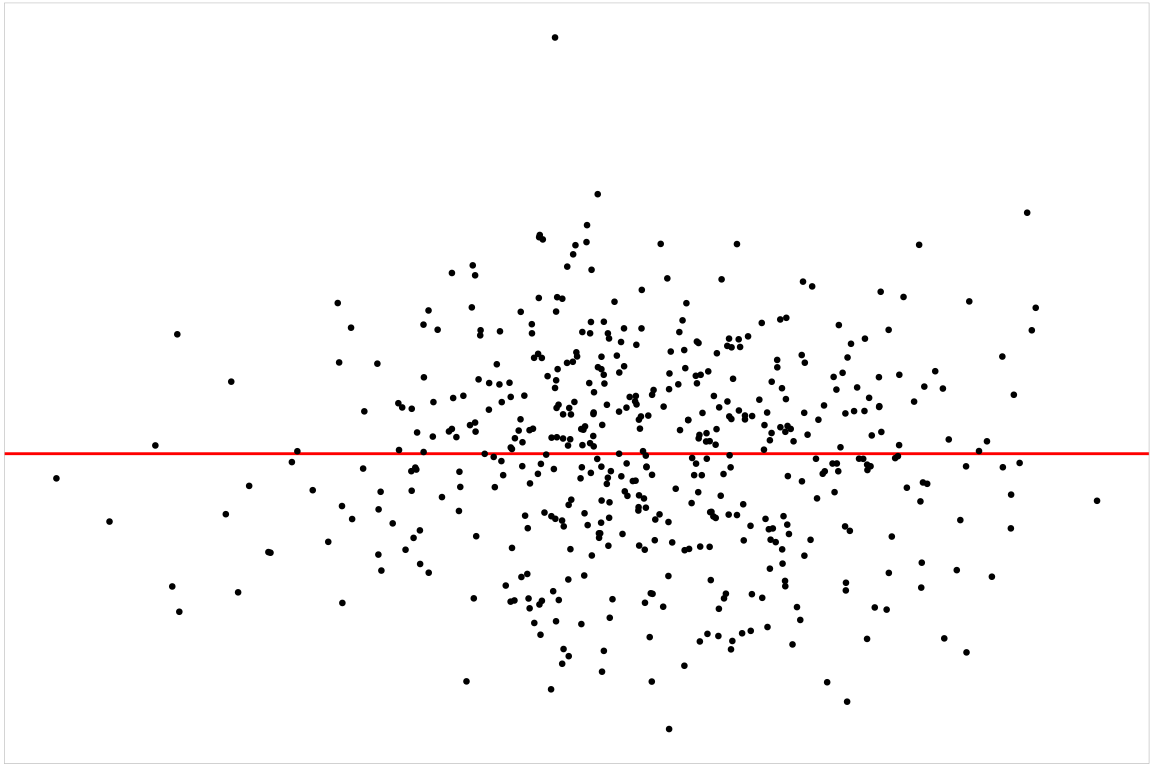
💡Example: Boston Housing
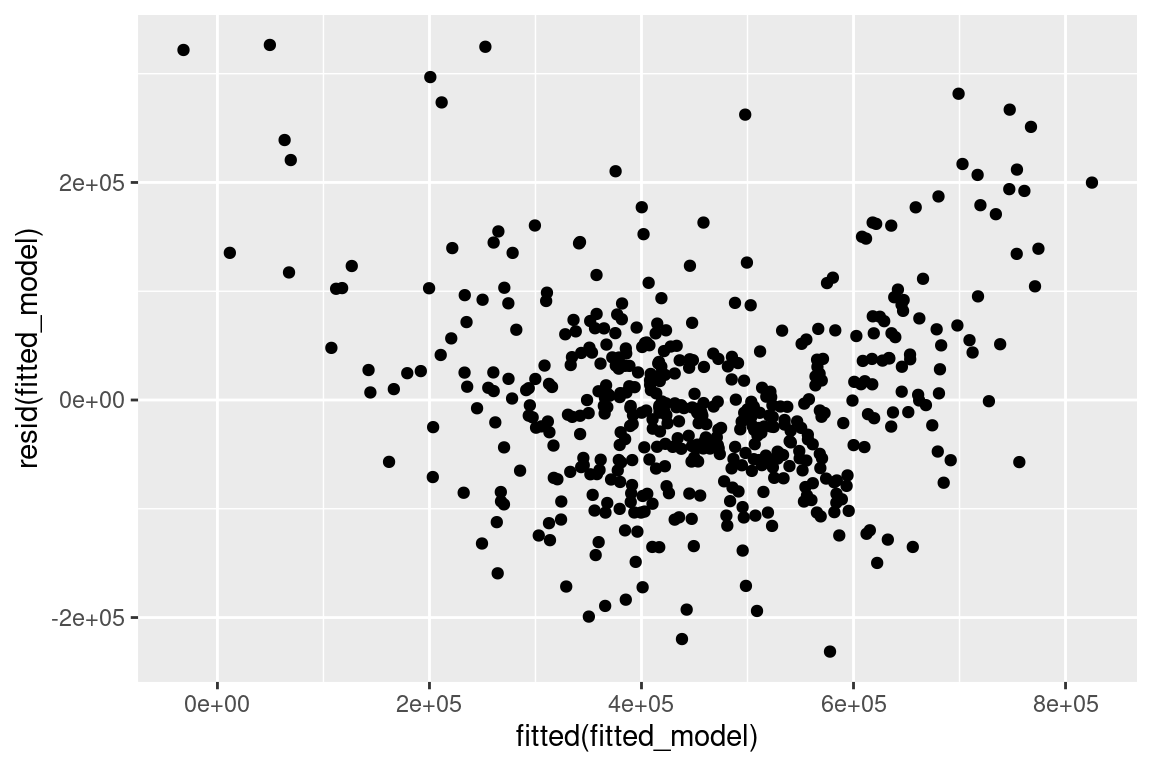
autovi💡Example: Boston Housing
Null residuals are simulated from the fitted model assuming it is correctly specified.
checker <- auto_vi(fitted_model = fitted_model,
keras_model = get_keras_model("vss_phn_32"))
checker$rotate_resid()# A tibble: 489 × 2
.fitted .resid
<dbl> <dbl>
1 632372. 24372.
2 525177. 13236.
3 646753. 54824.
4 624848. -98465.
5 611817. 188264.
6 551051. -67975.
7 504757. 142250.
8 445700. -175323.
9 281912. -101298.
10 453398. -121730.
# ℹ 479 more rows:::
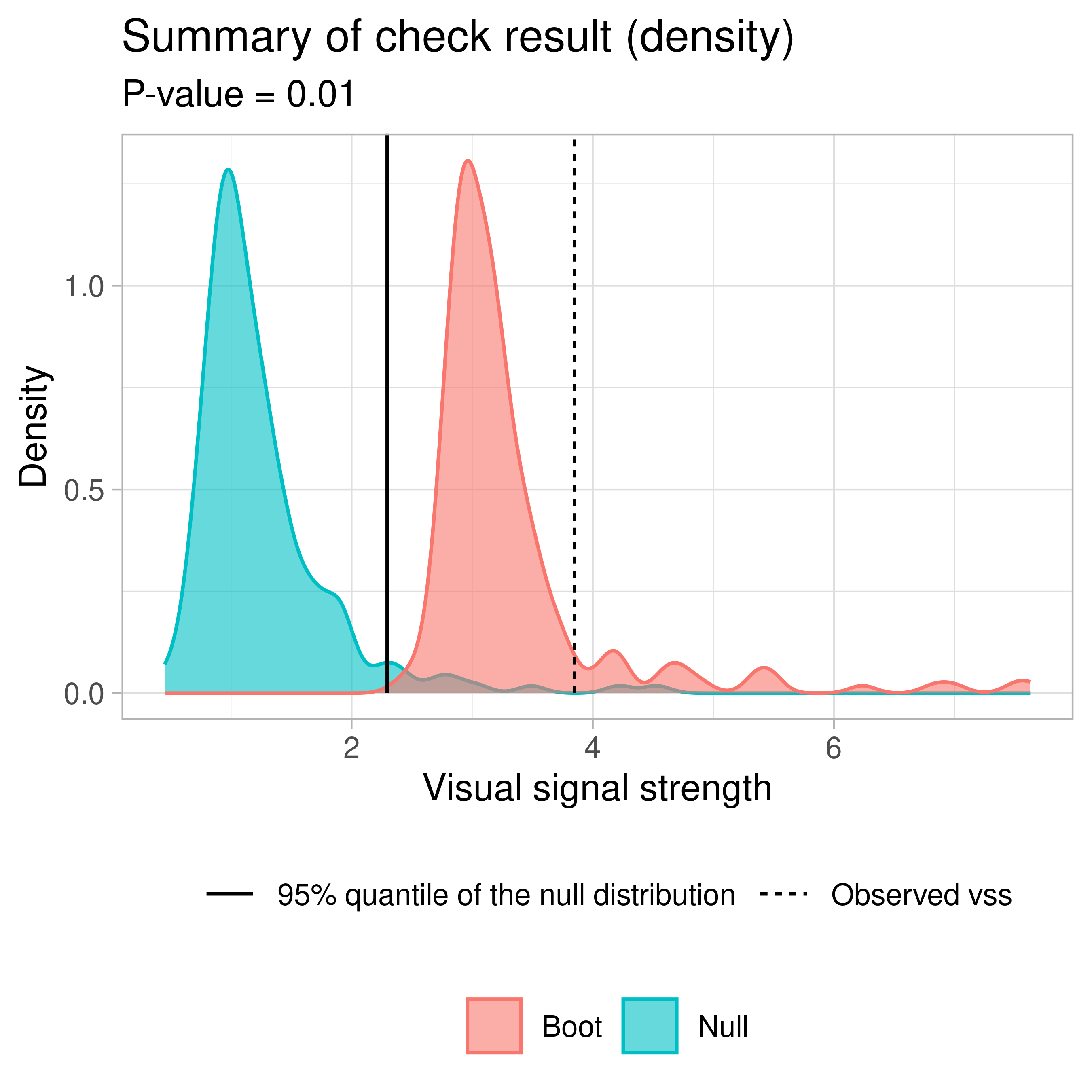
vss()
check()
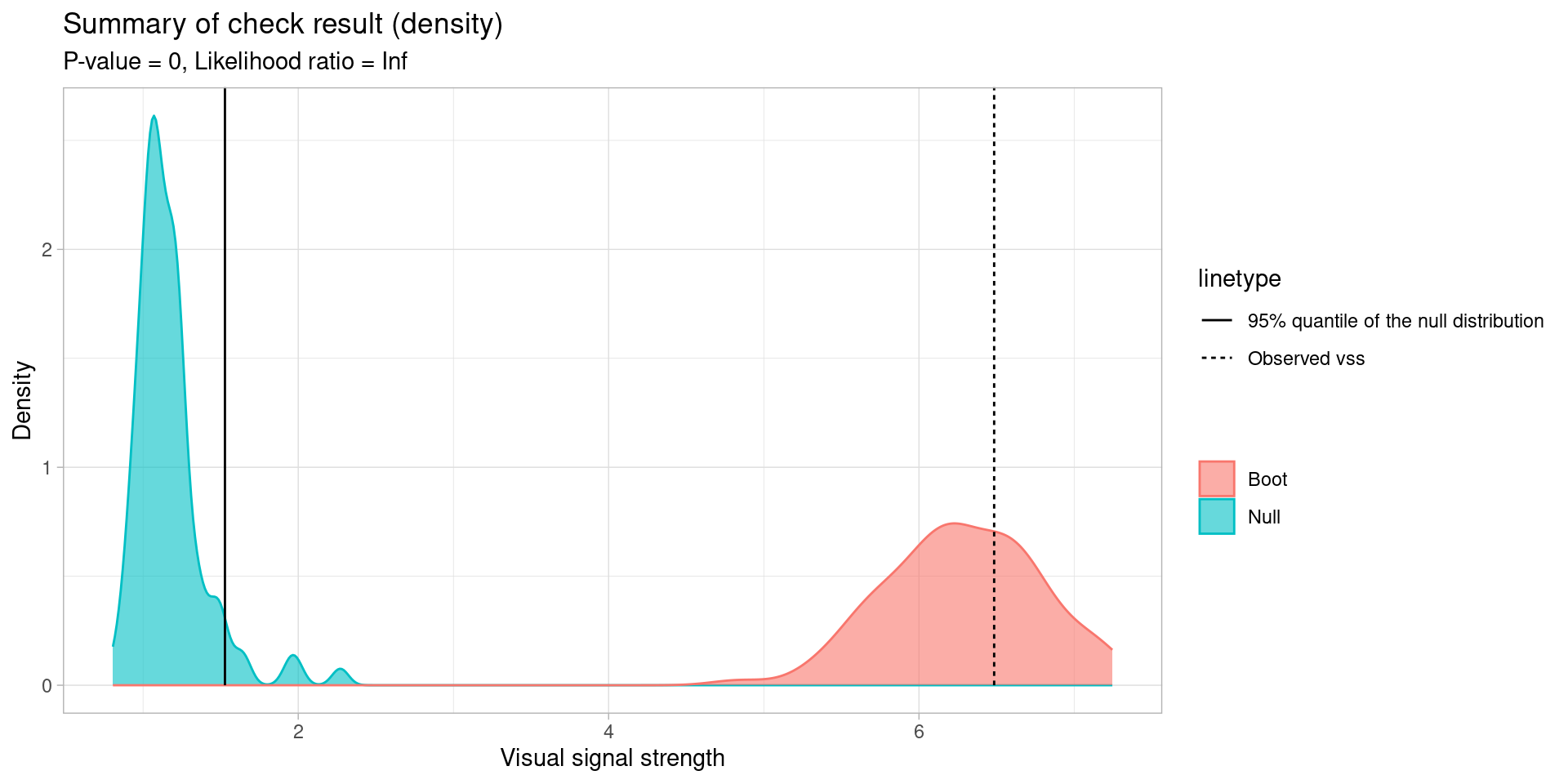
── <AUTO_VI object>
Status:
- Fitted model: lm
- Keras model: (None, 32, 32, 3) + (None, 5) -> (None, 1)
- Output node index: 1
- Result:
- Observed visual signal strength: 6.484 (p-value = 0)
- Null visual signal strength: [100 draws]
- Mean: 1.169
- Quantiles:
╔══════════════════════════════════════════╗
║ 25% 50% 75% 80% 90% 95% 99% ║
║1.037 1.120 1.231 1.247 1.421 1.528 1.993 ║
╚══════════════════════════════════════════╝
- Bootstrapped visual signal strength: [100 draws]
- Mean: 6.28 (p-value = 0)
- Quantiles:
╔══════════════════════════════════════════╗
║ 25% 50% 75% 80% 90% 95% 99% ║
║5.960 6.267 6.614 6.693 6.891 7.112 7.217 ║
╚══════════════════════════════════════════╝
- Likelihood ratio: 0.7064 (boot) / 0 (null) = Extremely large 💡Example: Dinosaur
RESET \(p\)-value = 0.742
B-P \(p\)-value = 0.36
S-W \(p\)-value = 9.21e-05
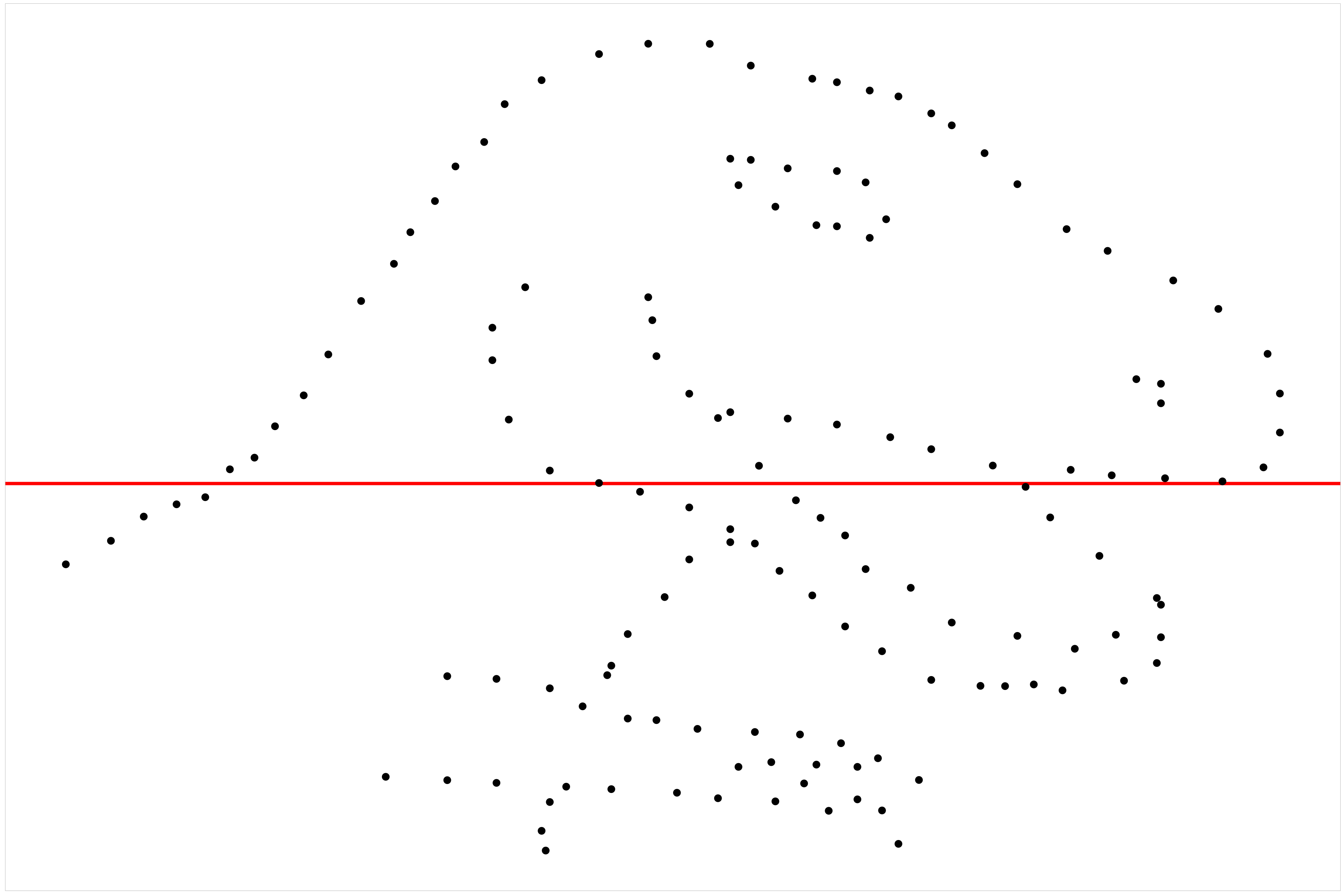
:::
🌐Shiny Application
Don’t want to install TensorFlow?
Try our shiny web application: https://shorturl.at/DNWzt
🧩Extensions
For GLMs and other regression models:
- Use raw residuals, but violations may not be identifiable and the test could be two-sided.
- Use transformed residuals that are roughly normally distributed.
- Reuse the pre-trained convolutional blocks and train a new computer vision model with an appropriate distance measure.
🎬Takeaway
You can use autovi to
Evaluate lineups of residual plots of linear regression models
Capture the magnitude of model violations through visual signal strength
Automatically detect model misspecification using a visual test
Thanks! Any questions?
Advisors