Take a look at the R markdown sample file I’ve created to go with the R script above. You can see the HTML file it generates here.
Download the Rmd file and open it with RStudio.
Change the output to output: word_document and hit the Render button  . Can you find the markdown-demo.docx file that was generated? What does it look like?
. Can you find the markdown-demo.docx file that was generated? What does it look like?
Change the output to output: pdf_document and hit the Render button  . Can you find the markdown-demo.pdf file that was generated? What does it look like?
. Can you find the markdown-demo.pdf file that was generated? What does it look like?
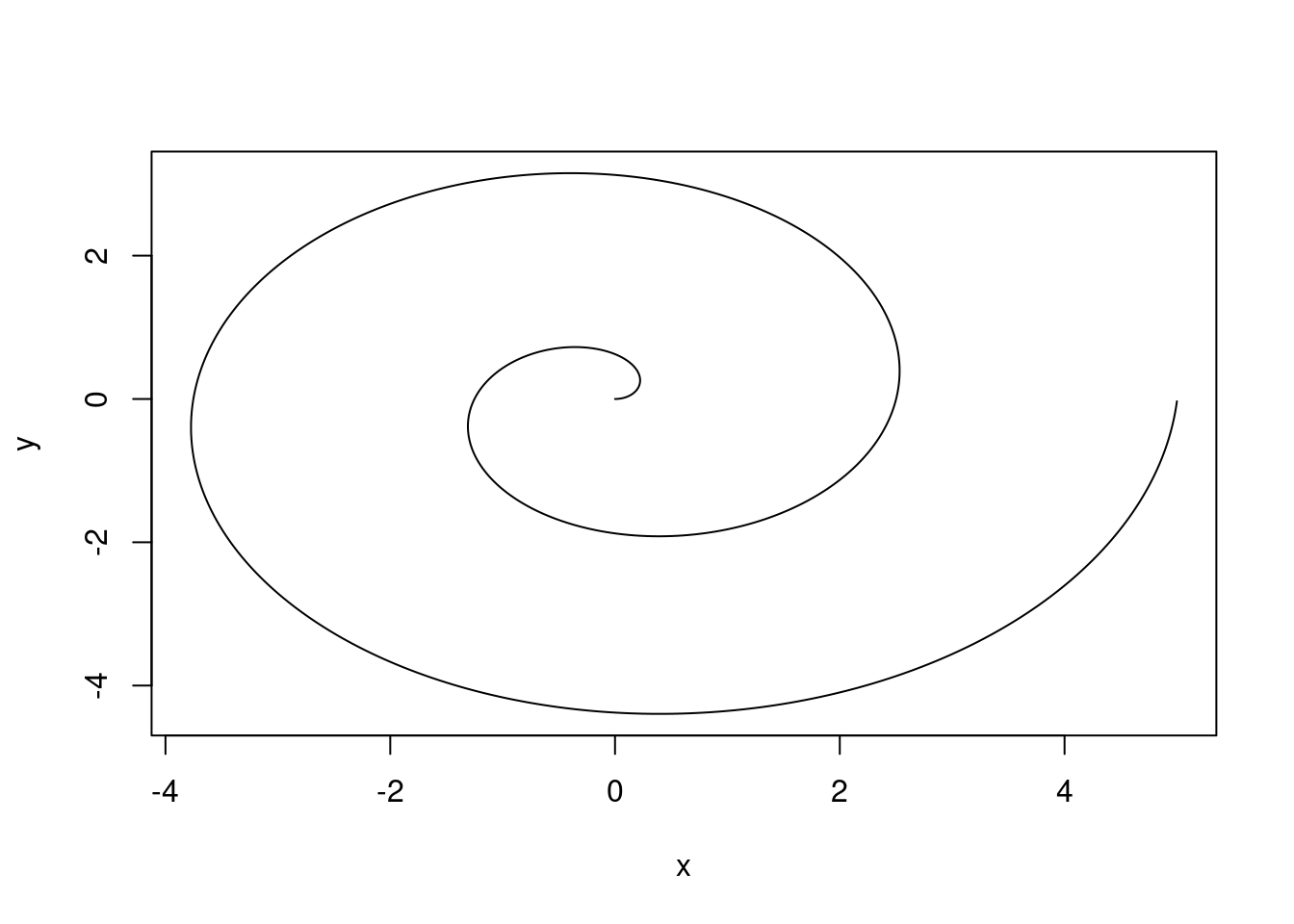
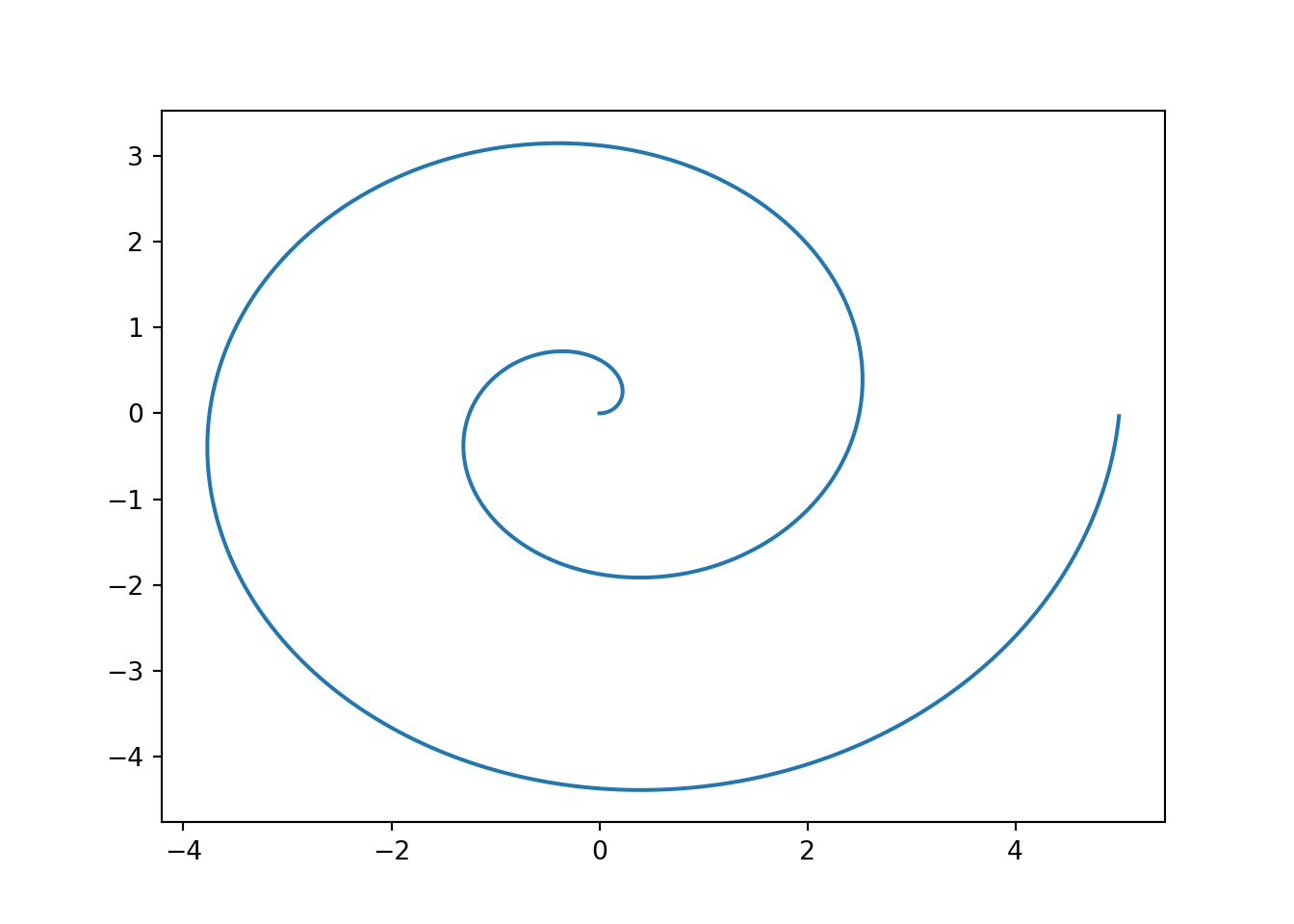
Rmarkdown tries very hard to preserve your formatted text appropriately regardless of the output document type. While things may not look exactly the same, the goal is to allow you to focus on the content and the formatting will “just work”.
Take a look at the jupyter notebook sample file I’ve created to go with the R script above. You can see the HTML file it generates here.
Download the ipynb file and open it with jupyter.
Export the notebook as a pdf file (File -> Save as -> PDF via HTML). Can you find the jupyter-demo.pdf file that was generated? What does it look like?
Export the notebook as an html file (File -> Save as -> HTML). Can you find the jupyter-demo.html file that was generated? What does it look like?
The nice thing about quarto is that it will work with python and R seamlessly, and you can compile the document using python or R. R markdown will also allow you to use python chunks, but you must compile the document in R.
Take a look at the Qmd notebook sample file I’ve created to go with the scripts above. You’ll notice that it is basically the script portion of this textbook – that’s because I’m writing the textbook in Quarto.
Download the qmd file and open it with RStudio
Try to compile the file by hitting the Render button 
(Advanced) In the terminal, type in quarto render path/to/file/quarto-demo.qmd. Does that render the HTML file? One advantage of this is that using quarto to render the file doesn’t require R at the command line. As the document contains R chunks, R is still required to compile the document, but the biggest difference between qmd and rmd is that qmd files are workflow agnostic - you can generate them in e.g. MS Visual Studio Code, compile them in that workflow, and never have to use RStudio.
 at the top of the editor window when the script is open.
at the top of the editor window when the script is open. . Can you find the markdown-demo.docx file that was generated? What does it look like?
. Can you find the markdown-demo.docx file that was generated? What does it look like?